Are we overlooking a critical factor in surgical site infections (SSIs)? In the quest for sterile operating rooms and antibiotic prophylaxis, a groundbreaking study suggests we've missed a vital piece of the puzzle: the patient's microbiome, a finding that challenges the wisdom of Semmelweis and may revolutionize infection prevention strategies.
It is inevitable when “cutting for cure” that the scalpel violates the integrity of our skin barriers while at the same time unleashing the contents of cells and cut blood vessels to create a veritable 24-hour all-you-can-eat buffet for bacteria. Surgical site infections (SSI) have been with us forever. It was Semmelweis, the “hand-washing” guy, that first focused our attention on the behavior of surgeons and their operating environment as the source of these infections rather than continuing to blame some environmental or social miasma.
The near-simultaneous development of sterile technique and garments, topical disinfectants, and antibiotics significantly reduced SSIs while spawning a set of preoperative “rituals” to ward off the bad jujus of SSI. There are no greater warriors against SSIs than orthopedic surgeons, in large part because their surgical procedures are very unforgiving of these infections. Bone, because of its relatively reduced blood supply, requires far more than a typical 7-10 day course of antibiotics to achieve a cure, and when the orthopedist implants artificial joints, they serve as “foreign bodies,” allowing infections places to fester and ultimately destroy the functional value of the newly repaired joint. [1]
Orthopedic surgical prophylaxis, the formal name for these rituals, includes protocols aimed at endogenous and exogenous sources of contamination:
- Testing and treating the patient’s nose for antibiotic-resistant bacteria.
- Various preoperative skin washing and disinfections that must be thorough but not too abrasive since injuring the skin creates another buffet for bacteria.
- Topical disinfection and systemic antibiotics at the moment of surgery.
- A range of alterations to the operating room environment to eliminate exogenous sources, including positive airflow that prevents outside air from entering the room, strict policies regarding who may or may not enter the room during the procedure, and encasing the surgical team in space suits so that they are less likely to contaminate the operative field and patient.
Additionally, the Center for Medicare Services (CMS) judges and grades hospitals on SSIs, and those hospitals doing the worst are economically penalized by reduced Medicare payments. Despite our best science and economic incentives, SSIs in “clean” surgery [2] remain stubbornly at 3-5% of cases. Is that the best we can do? Or was Dr. Semmelweis’s beliefs not entirely correct?
A new study suggests that the source of prophylaxis failures lies not in a lack of adherence to the rituals but with the patient’s microbiome. The dataset included 210 adult patients undergoing posterior spinal fusions, a “clean” surgery, who had “adequate preoperative nasal, rectal, and skin specimens” of their microbiome. These were not cultures; they were genomic signatures identifying various bacteria's presence. Fourteen patients, 6.8%, developed SSIs, and the genomic signatures of their cultured infections were compared to their microbiome signatures.
Genomic signatures are used to characterize our microbiome because they can identify the presence of microbes rather than their ability to infect. Amplification techniques can also provide information as to the relative abundance of one species over another. What these signatures do not tell us, especially for intestinal sampling, is where these bacteria are found anatomically – are they throughout the colon, or do they “hang out” around the cecum or perhaps the splenic flexure? This is all a preface to say that the study's first finding is that “an anatomic gradient exists along the skin microbiome of the back.”
Think of our skin as the home of our microbiome. Just as humans developed out of Africa and then spread across the Earth, the microbes of our mouth and nose (the oropharynx) form one “pole,” while the microbes of our “nether regions” form a more southern pole. From there, these very different bacteria spread out, with more of the oropharyngeal gathering in the skin overlying the spine of the neck and upper chest and those urogenital and rectal microbes overlying the sacral and lumbar spine’s skin.
“…stratification of microorganisms is a graded phenomenon with an inflection point occurring at about the mid-thoracic region …suggesting that it represents a common spatial anatomic feature of the human skin microbiome.”
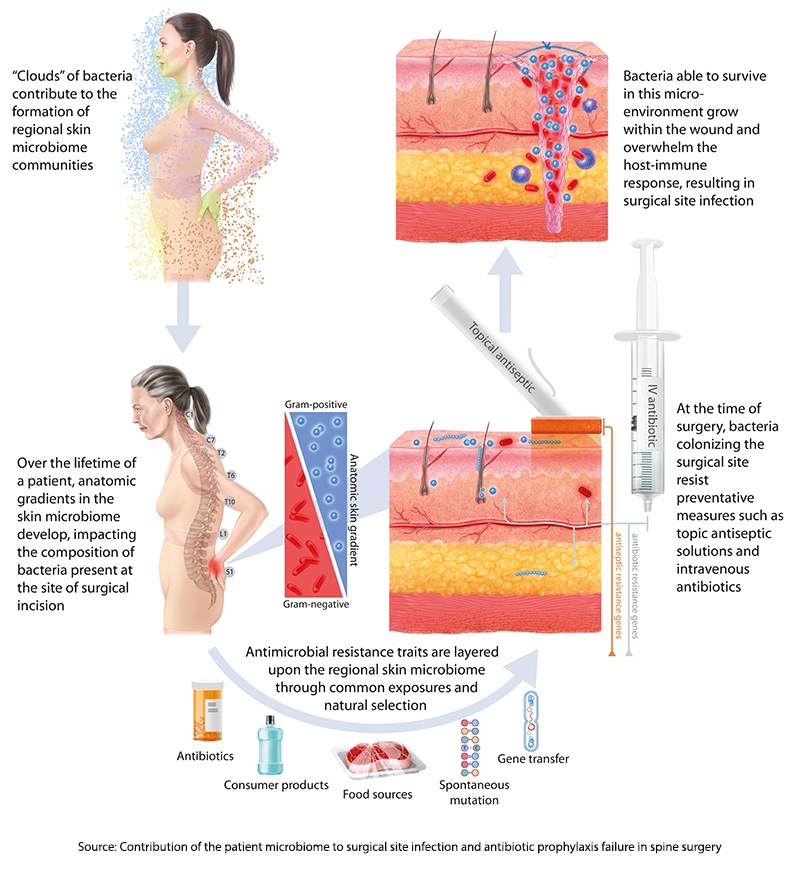 These skin microbes require very different antibiotics, so our current prophylaxis features broad-spectrum coverage. However, as it turns out, the researchers found that 86% of strains of bacteria already present on the patient’s skin before surgery had the same signature as those found in SSI cultures. Moreover, in roughly 75% of cases, these microbes followed that spatial anatomy [3] – more oropharyngeal sources of infection in cervical spine surgery and more urogenital and rectal microbes in lumbar spine surgery.
These skin microbes require very different antibiotics, so our current prophylaxis features broad-spectrum coverage. However, as it turns out, the researchers found that 86% of strains of bacteria already present on the patient’s skin before surgery had the same signature as those found in SSI cultures. Moreover, in roughly 75% of cases, these microbes followed that spatial anatomy [3] – more oropharyngeal sources of infection in cervical spine surgery and more urogenital and rectal microbes in lumbar spine surgery.
59% of the pathogens that caused SSIs were resistant to the antibiotics given at the time of surgery, indicating that bacteria with antibiotic-resistance genes were already circulating on the skin before the first cut. Genomic sequencing of “relevant genes” responsible for antimicrobial resistance found that these signatures could be identified pre-operatively 94% of the time from the preoperative samples of the patient’s microbiome. Finally, looking at a more extensive set of SSIs associated with these spinal procedures, there was no evidence that “exogenous strains,” those present in the environment, were responsible for any of the infections.
The bottom line is that preoperative genomic signatures can identify the most likely source of SSIs for a specific patient while also recognizing the presence of antimicrobial resistance. It may be possible to tailor antibiotic prophylaxis to the individual more precisely.
Semmelweis was only partially correct; the behavior and techniques of the surgeon and the patient’s microbiome both lead to SSIs. As the authors write,
“If these findings are replicated in other procedural cohorts, then this model of SSI pathogenesis could drive important shifts in infection prevention strategy and enable more individualized and patient-centered approaches. One potentially high-yield implication of this model is the personalized selection of surgical antibiotic prophylaxis. The marked individual differences in microbiome and resistome composition observed in this and most other human microbiome studies are not considered by current guidelines but could be routinely characterized before surgery. Such a strategy might guide a more sustainable framework that preserves the efficacy of surgical prophylaxis while limiting the trend of escalation to more broadly acting agents in the face of increasing population-level resistance.”
From Semmelweis's historical insights to modern genomic sequencing, our understanding of SSIs is evolving. By recognizing the influence of microbial composition and antibiotic resistance, there's potential for tailored prophylactic strategies. This paradigm shift promises a more personalized approach to surgical care and infection prevention.
[1] Surgeons operating on the large intestine are colorectal surgeons, who have rituals too. The surgical research literature continues to debate whether these surgical infections can be better prevented by pre-operatively emptying the colon of all stools, providing an antibiotic Armageddon to the colon lining, or combining the two.
[2] Clean surgery refers to procedures that do not involve body cavities or organs that contain microbes, e.g., the lungs, intestines, and urinary tract.
[3] This finding was based on a larger patient population of 1400 cases with culture-positive SSIs that could be correlated with the site of surgical intervention.
Source: Contribution of the patient microbiome to surgical site infection and antibiotic prophylaxis failure in spine surgery Science Translational Medicine DOI: 10.1126/scitranslmed.adk8222

