The sequence of an organism's genome, a staple in today's world of scientific experimentation, is as essential to scientific research as beakers. So, publishing over one thousand new bacterial genomes is like 'making it rain' to the microbiology research community.
An article entitled "1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life" was published this week in the journal Nature Biotechnology by an international research team led by the US Department of Energy's Joint Genome Institute. This release effectively doubles the number of currently available bacterial and archaeal genomes available to researchers currently.
This work is part of the Genomic Encyclopedia of Bacteria and Archaea Initiative (GEBA-I), which was founded by Jonathan Eisen, Ph.D. Eisen's website aims to "expand the reference genome catalog of broad phylogenetic and physiological diversity, to determine how this catalog facilitates the discovery of protein families and expands the diversity of known functions, and to ascertain whether these type-strain genomes improve the recruitment and phylogenetic assignment of existing metagenomic sequences."
To get a sense of how much information has been added to the collective database of microbial genomes, see Figure 1 (below.) A refresher in the taxonomic classification of animals is given below as a 'note'. (1)
Each spoke of the wheel in Figure 1 represents one phylum (a large group of organisms that are alike.) To give this particular taxonomic grouping some perspective, the phylum for humans is 'chordata', which includes 65,000 animals, including all vertebrates. The phyla that contain a GEBA-1 genome are denoted by a triangle along the arm that is colored red (24 arms out of 40.) The phyla without a GEBA-1 genome are marked by grey triangles. At the end of the arms are circular pie charts to indicate how many new genomes in that phyla are from the GEBA-1 data (red) compared to the total number of known sequences per phylum (shown in blue.) The number at the end of the arms represents the total number of new genera (plural of genus) added to that phylum by the GEBA-1 data. The bottom line, without getting too bogged down, is that the information from the GEBA-1 data has provided a ton of new information to the already existing bacterial 'tree of life."
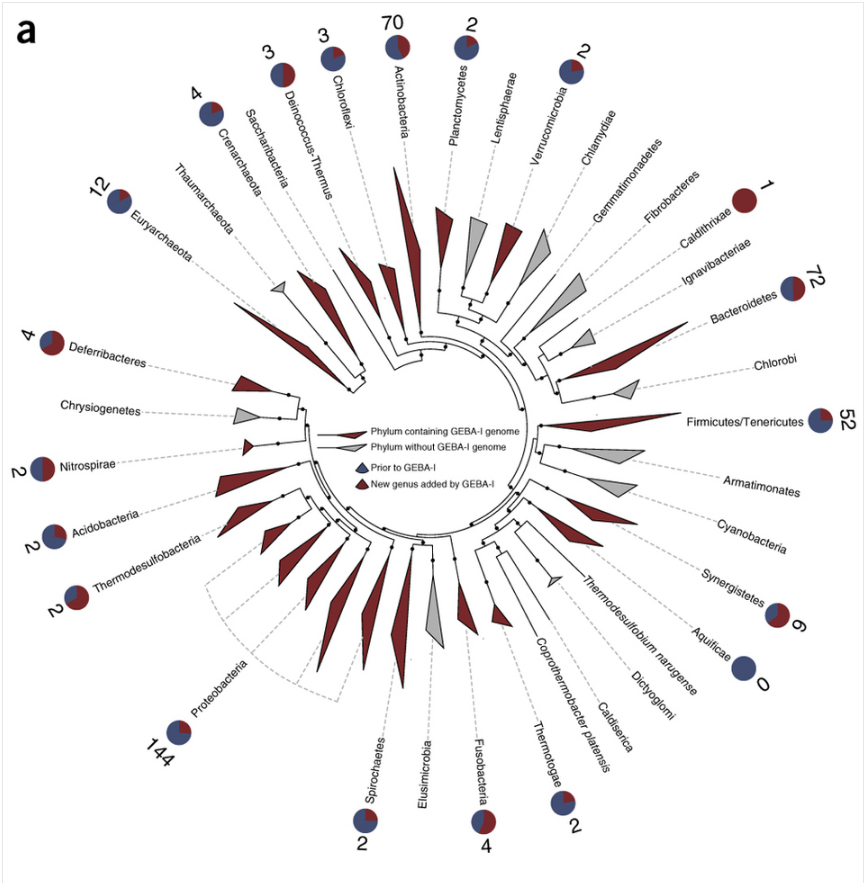
Figure 1 - GEBA-I strain phylogeny and distribution.
Since sequencing has become so fast and cheap, this may not seem like huge news. However, bacterial genomes are generally only focused upon when they have clinical significance. In fact, in 2015, 43 percent of the bacterial genome sequences that were available came from ten bacterial species, all of which are pathogenic to humans. Given the amount of bacterial diversity that exists, this resulted in highly skewed and unbalanced information with many knowledge gaps.
The GEBA-1 team identified gaps by analyzing the known genomes compiled in the All-Species Living Tree Project. The team then isolated specimens and sequenced them using Illumina sequencing.
Now we have a huge amount of new information becoming available, including previously unassigned proteins and gene clusters.
The researchers wrote, "this resource data set is the single largest effort (to our knowledge) to increase the phylogenetic coverage of cultivated bacterial and archaeal isolates." This paper marks the largest ever release of new genomes. For the scientific community, it is a great tool and will certainly open up new avenues for investigation into different avenues of study on bacterial diversity.
NOTES:
(1) Taxonomic Names