"Every city has its own 'molecular echo' of the microbes that define it.”
Christopher Mason, Ph.D., Director of the WorldQuant Initiative for Quantitative Prediction
One of the early COVID-19 lessons was that cities, and our “built environment” in general, could affect our health – consider the population density of low-income housing or a subway car at rush hour and its impact of exposure to COVID-19. Advances in DNA analysis have allowed statistical methods to describe the microbes present in a microbiome without cultures. While that introduces a bit of ambiguity [1], it does allow for descriptions that we might not make because of the limitations in our culturing bacteria, viruses, and fungi. The study of geographic microbiomes at this scale is in its infancy; none of this is settled science, so the research paints with an admittedly large, and at times, undiscerning brush.
The researchers took 4,728 samples from 60 urban mass transit systems globally, from three common surfaces, railings, benches, and ticket kiosks – the transit systems “stood-in” for a particular city’s built environment. The samples under statistical analysis, more properly shotgun metagenomic sequencing, to describe the various microbes present.
- Thirty-one species were “core” taxa [2] found in 97% of urban environments. Their relative abundance changed from city to city
- 1,425 species reflected sub-core taxa found in 70% of more of the cities.
- Several species remained unidentified by their methods, but the researchers believe that they were able to identify about 80%
- At least based upon its metagenomics, the microbiome's diversity was greater at the equator than at higher latitudes. Samples were taken from surfaces we handle like railings or doorknobs had microbiomes more closely aligned with human skin’s microbial patterns. That said, climate seemed more “determinate” than human interaction.
- Sampling done one year apart showed changes in an urban microbiome; it is not static but evolving; although these differences were not as significant as those between cities in any given year.
- City climate was the strongest driver of diversity. Microbiomial patterns were not affected by population density.
- Taken together and using machine learning, researchers could successfully identify the geographic source of a sample 88% of the time
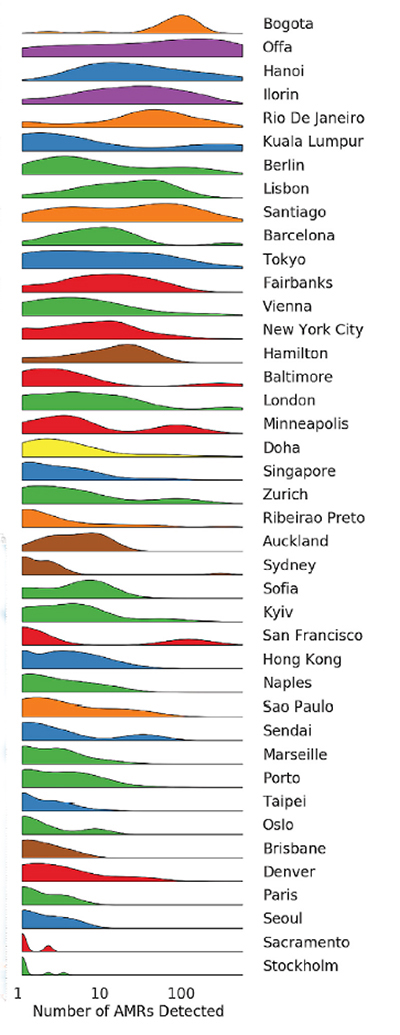 Markers of antimicrobial resistance (AMR) were widespread but not necessarily abundant and showed some degree of “geographic specificity.” Some forms of antimicrobial resistance were more common in some cities than others. There was a “city signature” to AMR but not as strong as the microbial pattern – researchers could accurately predict a sample’s geographic source 77% of the time from the AMR.
Markers of antimicrobial resistance (AMR) were widespread but not necessarily abundant and showed some degree of “geographic specificity.” Some forms of antimicrobial resistance were more common in some cities than others. There was a “city signature” to AMR but not as strong as the microbial pattern – researchers could accurately predict a sample’s geographic source 77% of the time from the AMR. - The preponderance of AMR globally impacted the macrolide, lincosamide, and streptogramin or MLS family of antimicrobials and the beta-lactams, all of which are important in treating a wide variety of human illness.
While the study raises more questions than it answers, as good research should, we can make a few generalizations. First, our urban built environments create microbial niches. The relative contribution of drivers like urban density, housing, design is unknown, but it seems at first blush that climate, as determined by latitude, plays an important role. Second, these microbiological ecologies are subject to continuous evolution; they are not static. Third, while we can see the impact of our behavior, as measured by antimicrobial resistance, it is clear from the geographical diversity of those effects that other factors are at play on the urban microbiome. These are the unknown unknowns waiting to be explored.
Nature remains both awe-inspiring and enigmatic.
[1] One of the study’s limitations, as the researchers point out, is that a large number of genetic samples that could not be identified
[2] Taxa is a grouping by a biological definition
Source: A global metagenomic map of urban microbiomes and antimicrobial resistance Cell DOI: 10.1016/j.cell.2021.05.002




